
ChordexBio is advancing genomic intelligence through FAS2rDNA
Automated and scalable DNA reconstruction for AI-driven genomics
DNA sequence reconstruction | genomic automation | AI-driven genomics | machine learning sequence analysis
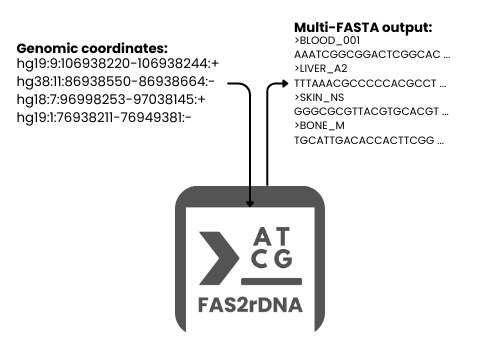
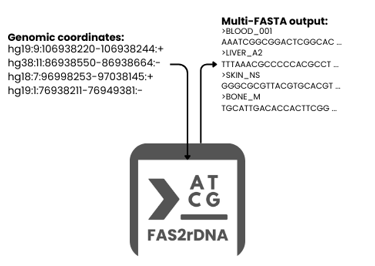
In the era of big data, genomic information holds the key to understanding life, disease, and innovation. Yet, extracting actionable sequences from raw genomic coordinates, especially across large, complex datasets can be slow, error-prone, and resource-intensive. FAS2rDNA is changing that. We developed FAS2rDNA to provide researchers with automated, reliable, and scalable DNA reconstruction that directly address workflow gaps but also supports AI and machine learning applications.
Unlocking the value of genomic data
FAS2rDNA transforms raw genomic coordinates into ready-to-use DNA sequences, giving scientists the ability to:
Accelerate research workflows by eliminating manual sequence extraction
Ensure reproducibility and accuracy across studies and datasets
Power AI and ML models with high-quality, standardized inputs
By streamlining these steps, FAS2rDNA turns what was once a technical bottleneck into a launchpad for genomic intelligence, enabling researchers to focus on discovery rather than data wrangling.
--- page break ---
Driving AI-enabled insights
Machine learning thrives on consistent, high-quality data. FAS2rDNA delivers precisely that, supporting predictive genomics, isoform-level analyses, and pan-cancer studies like miRNome reconstruction in TCGA cohorts. Researchers can now feed AI pipelines with well-structured, reproducible sequences, improving model accuracy, interpretability, and discovery potential.
Learn more about the use cases of FAS2rDNA here:
High-throughput isoform-wide miRNome sequence reconstruction in the TCGA-LUAD cohort using FAS2rDNA (https://dx.doi.org/10.17504/protocols.io.rm7vzenqxvx1/v1)
FAS2rDNA-Colab: A cloud-based workflow for pan-cancer, isoform-wide miRNome reconstitution across TCGA cohorts (https://dx.doi.org/10.17504/protocols.io.14egn1xr6v5d/v1)
Accessible and scalable
Whether used through the command-line interface for large-scale pipelines or the Colab implementation for rapid exploration, FAS2rDNA is designed to meet the needs of modern genomic research. Its flexibility ensures that both small teams and large collaborative projects can harness its power without technical friction. Learn more about FAS2rDNA versions here:
FAS2rDNA-CLI: https://github.com/mahvin92/FAS2rDNA
FAS2rDNA-Colab: https://github.com/mahvin92/FAS2rDNA-Colab
Why it matters
Every nucleotide matters. FAS2rDNA amplifies the value hidden in genomic coordinates, bridging the gap between raw data and actionable insights. By enabling AI-driven analysis at scale, it is helping researchers advance genomic intelligence, accelerating discoveries that impact medicine, biotechnology, and fundamental biology.
We value your feedback, tell us what you think. Contact us
Related stories:
ChordexBio is advancing genomic intelligence through FAS2rDNA
Discover FAS2rDNA, a smart tool that automates DNA sequence reconstruction from genomic coordinates, providing reproducible sequences for AI and machine learning-driven genomic analysis. Ideal for large-scale and multi-assembly studies.
BLOGFAS2RDNA
12/30/20252 min read
Algorithms empowering lives
© 2026 CHORDEXBIO RESEARCH.
All rights reserved.
Engage with us:


